Difference between revisions of "Antimony"
(→Features overview) |
(→Features overview) |
||
| Line 56: | Line 56: | ||
<td> | <td> | ||
<ul> | <ul> | ||
| − | <li>Can import only | + | <li>Can import only diagram from the current collection,</li> |
<li>The public port is created only if it is registered in a parameters model,</li> | <li>The public port is created only if it is registered in a parameters model,</li> | ||
<li>Some declarations don't supported: <i> DNA strand definitions, deletions</i>,</li> | <li>Some declarations don't supported: <i> DNA strand definitions, deletions</i>,</li> | ||
Revision as of 16:36, 31 October 2013
Contents |
Antimony plugin
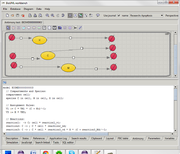
Antimony plugin for BioUML 0.9.6 aims to combine two representations of mathematical model (particularly - SBML model):
- visual representation as BioUML-diagram using extended SBGN notation
- text-based representation using Antimony language[1].
Plugin allows user to edit model both visually and as text.
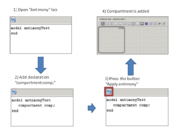
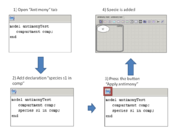
If visual repesentation is changed - antimony text is updated on the fly.
Text changes are applied to the diagram on demand (after pressing "Apply Antimony" button).
Besides model representations syncrhonisation, we also try to presereve user diagram layout as well as antimony text format (spaces, comments, etc.)
Please note: antimony changes will be applied to the diagram only after "Apply Antimony" button is pressed.
If you change diagram before you press this button, all changes in text will be lost!
Features overview
Currently plugin is in beta version, some features of antimony aren't supported. Also we want to achieve full synchronization between antimony and sbml-model in BioUML. And for this purpose we need some additions for the antimony specification.
| Are supported within an antimony | Necessary additions in the antimony specification |
|
|
| Antimony features which hadn't supported in beta-version | |
|
References
- ↑ Smith, L.P., Bergmann, F.T., Chandran, D. Sauro, M.H. Antimony: a modular model definition language. Bioinformatics, 2009, 25(18): 2452-2454. doi:10.1093/bioinformatics/btp401