Difference between revisions of "BioBlend"
(Created page with "BioBlend is a unified API in Python that wraps the functionality of Galaxy and CloudMan APIs <cite>Sloggett2013</cite>. Automated installation of BioBlend is available v...") |
|||
| (3 intermediate revisions by one user not shown) | |||
| Line 1: | Line 1: | ||
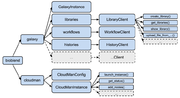
| + | [[File:BioBlend.png|thumb|BioBlend structure]] | ||
BioBlend is a unified API in Python that wraps the functionality of Galaxy and [[CloudMan]] APIs <cite>Sloggett2013</cite>. | BioBlend is a unified API in Python that wraps the functionality of Galaxy and [[CloudMan]] APIs <cite>Sloggett2013</cite>. | ||
| Line 6: | Line 7: | ||
There is Java version blend4j, but it supports only Galaxy API. | There is Java version blend4j, but it supports only Galaxy API. | ||
| − | https://github.com/jmchilton/ | + | https://github.com/jmchilton/blend4j |
Links: | Links: | ||
*documentation - http://bioblend.readthedocs.org | *documentation - http://bioblend.readthedocs.org | ||
*presentation - https://s3.amazonaws.com/usecloudman/BOSC2013_BioBlend.pdf | *presentation - https://s3.amazonaws.com/usecloudman/BOSC2013_BioBlend.pdf | ||
| − | *source code - https://github.com/afgane/bioblend | + | *source code - https://github.com/afgane/bioblend (43 files, ~200 kb). |
== CloudMan examples== | == CloudMan examples== | ||
| Line 63: | Line 64: | ||
#Sloggett2013 pmid=23630176 | #Sloggett2013 pmid=23630176 | ||
</biblio> | </biblio> | ||
| − | + | http://bib.irb.hr/datoteka/627609.PUBLISHED_BioBlend.pdf | |
[[Category:Galaxy]] | [[Category:Galaxy]] | ||
[[Category:Cloud]] | [[Category:Cloud]] | ||
Latest revision as of 21:57, 5 January 2014
BioBlend is a unified API in Python that wraps the functionality of Galaxy and CloudMan APIs [1].
Automated installation of BioBlend is available via PyPI (e.g. pip install bioblend).
The library has been tested and is working on Linux, Macintosh and Windows-based systems.
There is Java version blend4j, but it supports only Galaxy API. https://github.com/jmchilton/blend4j
Links:
- documentation - http://bioblend.readthedocs.org
- presentation - https://s3.amazonaws.com/usecloudman/BOSC2013_BioBlend.pdf
- source code - https://github.com/afgane/bioblend (43 files, ~200 kb).
[edit] CloudMan examples
Create a CloudMan compute cluster, via an API and directly from your local machine:
from bioblend.cloudman import CloudManConfig
from bioblend.cloudman import CloudManInstance
cfg = CloudManConfig('<your cloud access key>', '<your cloud secret key>', 'My CloudMan', 'ami-<ID>', 'm1.small', '<password>')
cmi = CloudManInstance.launch_instance(cfg)
cmi.get_status()
Reconnect to an existing CloudMan instance and manipulate it:
from bioblend.cloudman import CloudManInstance
cmi = CloudManInstance("<instance IP>", "<password>")
cmi.add_nodes(3)
cluster_status = cmi.get_status()
cmi.remove_nodes(2)
[edit] Implementation details
BioBlend uses REST protocol to Interact with Galaxy and CloudMan.
For example,
REST call: http://main.g2.bx.psu.edu/api/histories/0a7b7992a7cabaec?key=123456789abcdef10
Returns JSON:
{ 'id': '0a7b7992a7cabaec',
'name': 'Output history',
'state_details': { 'discarded': 0,
'empty': 0, ......
In BioBlend it becomes Python:
GalaxyInstance.histories.show_history('0a7b7992a7cabaec')
Returns corresponding Python dictionary
{ 'id': '0a7b7992a7cabaec',
'name': 'Output history',
'state_details': { 'discarded': 0,
'empty': 0, .....
[edit] References
- Error fetching PMID 23630176: