Difference between revisions of "Mapping to ontologies (Gene table) (workflow)"
(GeneXplain -> geneXplain) |
(Automatic synchronization with BioUML) |
||
| (3 intermediate revisions by one user not shown) | |||
| Line 6: | Line 6: | ||
[[File:Mapping-to-ontologies-Gene-table-workflow-overview.png|400px]] | [[File:Mapping-to-ontologies-Gene-table-workflow-overview.png|400px]] | ||
== Description == | == Description == | ||
| − | + | This workflow is designed to classify an input gene set to several ontologies and to identify terms, hits for which are overrepresented in the input set. | |
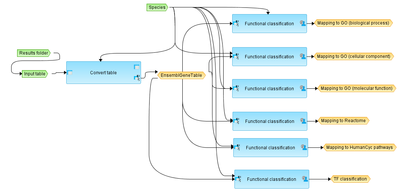
| − | + | The input file can be any gene or protein table. | |
| − | This table with Ensembl Gene Ids is subjected to functional classification, which is done in parallel by the following ontologies: GO biological processes, GO cellular components, GO molecular functions, Reactome pathways, HumanCyc pathways, TF classification. | + | At the first step, the input table is converted into a table with Ensembl Gene IDs. This table with Ensembl Gene Ids is subjected to functional classification, which is done in parallel by the following ontologies: GO biological processes, GO cellular components, GO molecular functions, Reactome pathways, HumanCyc pathways, TF classification. |
For each ontological term several parameters are calculated, including expected number of hits, actual number of hits, p-value, as well as hit names and the link to the corresponding ontological term. | For each ontological term several parameters are calculated, including expected number of hits, actual number of hits, p-value, as well as hit names and the link to the corresponding ontological term. | ||
| Line 20: | Line 20: | ||
[[Category:Workflows]] | [[Category:Workflows]] | ||
| + | [[Category:GeneXplain workflows]] | ||
[[Category:Autogenerated pages]] | [[Category:Autogenerated pages]] | ||
Latest revision as of 16:34, 12 March 2019
- Workflow title
- Mapping to ontologies (Gene table)
- Provider
- geneXplain GmbH
[edit] Workflow overview
[edit] Description
This workflow is designed to classify an input gene set to several ontologies and to identify terms, hits for which are overrepresented in the input set.
The input file can be any gene or protein table.
At the first step, the input table is converted into a table with Ensembl Gene IDs. This table with Ensembl Gene Ids is subjected to functional classification, which is done in parallel by the following ontologies: GO biological processes, GO cellular components, GO molecular functions, Reactome pathways, HumanCyc pathways, TF classification.
For each ontological term several parameters are calculated, including expected number of hits, actual number of hits, p-value, as well as hit names and the link to the corresponding ontological term.
[edit] Parameters
- Input table
- Species
- Results folder