Difference between revisions of "Gene set enrichment analysis (Gene table) (workflow)"
(geneXplain -> GeneXplain in category name) |
m (Cosmetic changes in category formatting) |
||
| Line 22: | Line 22: | ||
[[Category:Workflows]] | [[Category:Workflows]] | ||
| − | |||
[[Category:GeneXplain workflows]] | [[Category:GeneXplain workflows]] | ||
| + | [[Category:Autogenerated pages]] | ||
Revision as of 13:28, 16 May 2013
- Workflow title
- Gene set enrichment analysis (Gene table)
- Provider
- geneXplain GmbH
Workflow overview
Description
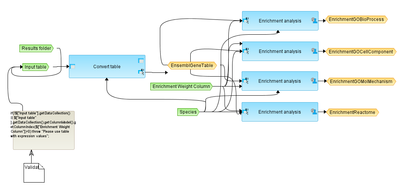
This workflow is designed to perform Gene Set Enrichment Analysis, GSEA, as it is described at http://www.broadinstitute.org/gsea/index.jsp. As input, any gene or protein table that includes fold change (or any other numerical value that can be used as a weighted column) can be taken.
At the first step, the input table is converted into a table with Ensembl Gene IDs.
This table with Ensembl Gene IDs is subjected to GSEA. Enrichment analysis is done in parallel by the following ontologies: GO biological processes, GO cellular components, GO molecular functions and by the Reactome pathways.
For each ontological term several parameters are calculated, including nominal p-value, ES, NES, as well as hit names, the link to the corresponding ontological term, and the link to open a visualization plot.
Parameters
- Input table
- Enrichment Weight Column
- Enter the name of the column from the Input table with numerical values
- Species
- Results folder