Difference between revisions of "Systems biology - model simulation"
From BioUML platform
Ilya Kiselev (Talk | contribs) |
|||
| Line 1: | Line 1: | ||
<font size=3> | <font size=3> | ||
| − | |||
To simulate SBML model, do the following: | To simulate SBML model, do the following: | ||
| − | |||
<ol> | <ol> | ||
| − | <li> Go to the '''Simulation''' tab in the right bottom panel. Depending on the type of your diagram, the appropriate simulation engine will be set in the '''Selected engine''' field. For example, '''ODE Simulation Engine''' is used for ordinary differential equation models. For more information on supported simulation engines, see the [[Simulation]] section. | + | <li> Go to the '''Simulation''' tab in the right bottom panel. It has two inner tabs: '''Engine''' and '''Plot'''. First we will need to set variables to plot. Got to '''Plot''' tab and set value for default curve. |
| + | |||
| + | <br><br>[[File:systems_biology_simulation_0.png]]<br><br> | ||
| + | |||
| + | <li> Next step is to go to '''Engine''' tab and set simulation properties. | ||
| + | Depending on the type of your diagram, the appropriate simulation engine will be set in the '''Selected engine''' field. For example, '''ODE Simulation Engine''' is used for ordinary differential equation models. For more information on supported simulation engines, see the [[Simulation]] section. | ||
| − | <br><br>[[File: | + | <br><br>[[File:systems_biology_simulation_1.png]]<br><br> |
<li> Select one of the available simulators or use the default one: | <li> Select one of the available simulators or use the default one: | ||
| Line 16: | Line 19: | ||
*[[JVODE|JVode]] (default) | *[[JVODE|JVode]] (default) | ||
| − | <br>[[File: | + | <br>[[File:systems_biology_simulation_2.png]]<br><br> |
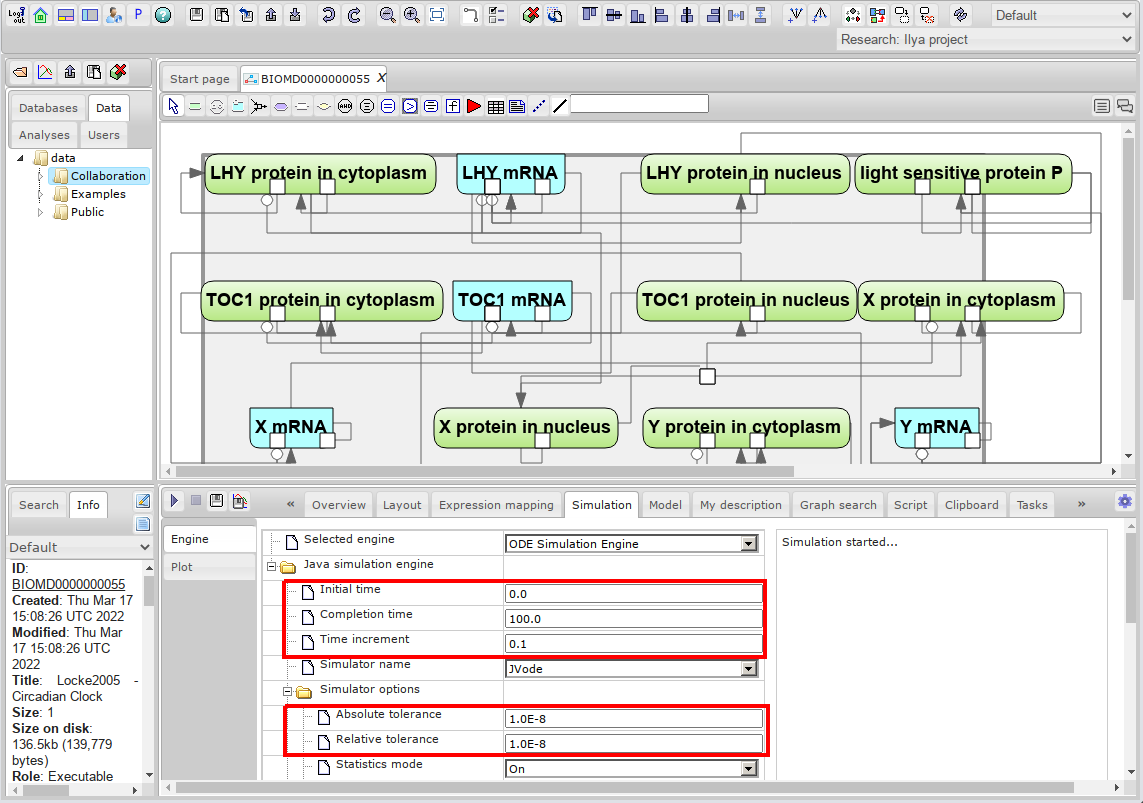
<li> Set simulation options: | <li> Set simulation options: | ||
| Line 23: | Line 26: | ||
*'''Time increment''' - the gap between the time points at which the simulation results will be written. | *'''Time increment''' - the gap between the time points at which the simulation results will be written. | ||
| − | <br>[[File: | + | <br>[[File:systems_biology_simulation_3.png]]<br><br> |
<li>Click on the '''Simulate''' button [[File:Execute_icon.png]]. The window that opens will show the results of simulation of the variables selected to plot. | <li>Click on the '''Simulate''' button [[File:Execute_icon.png]]. The window that opens will show the results of simulation of the variables selected to plot. | ||
| − | <br><br>[[File: | + | <br><br>[[File:systems_biology_simulation_4.png]]<br><br> |
</ol> | </ol> | ||
</font> | </font> | ||
Revision as of 14:59, 18 March 2022
To simulate SBML model, do the following:
- Go to the Simulation tab in the right bottom panel. It has two inner tabs: Engine and Plot. First we will need to set variables to plot. Got to Plot tab and set value for default curve.
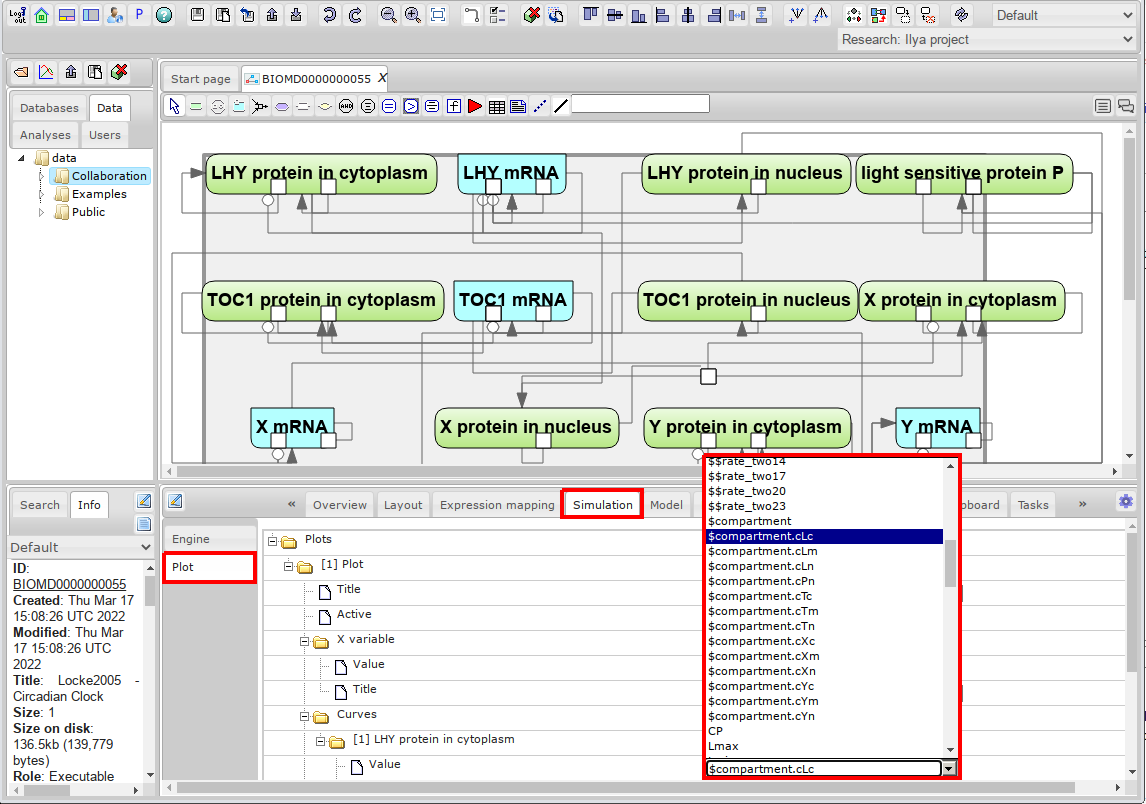
- Next step is to go to Engine tab and set simulation properties.
Depending on the type of your diagram, the appropriate simulation engine will be set in the Selected engine field. For example, ODE Simulation Engine is used for ordinary differential equation models. For more information on supported simulation engines, see the Simulation section.
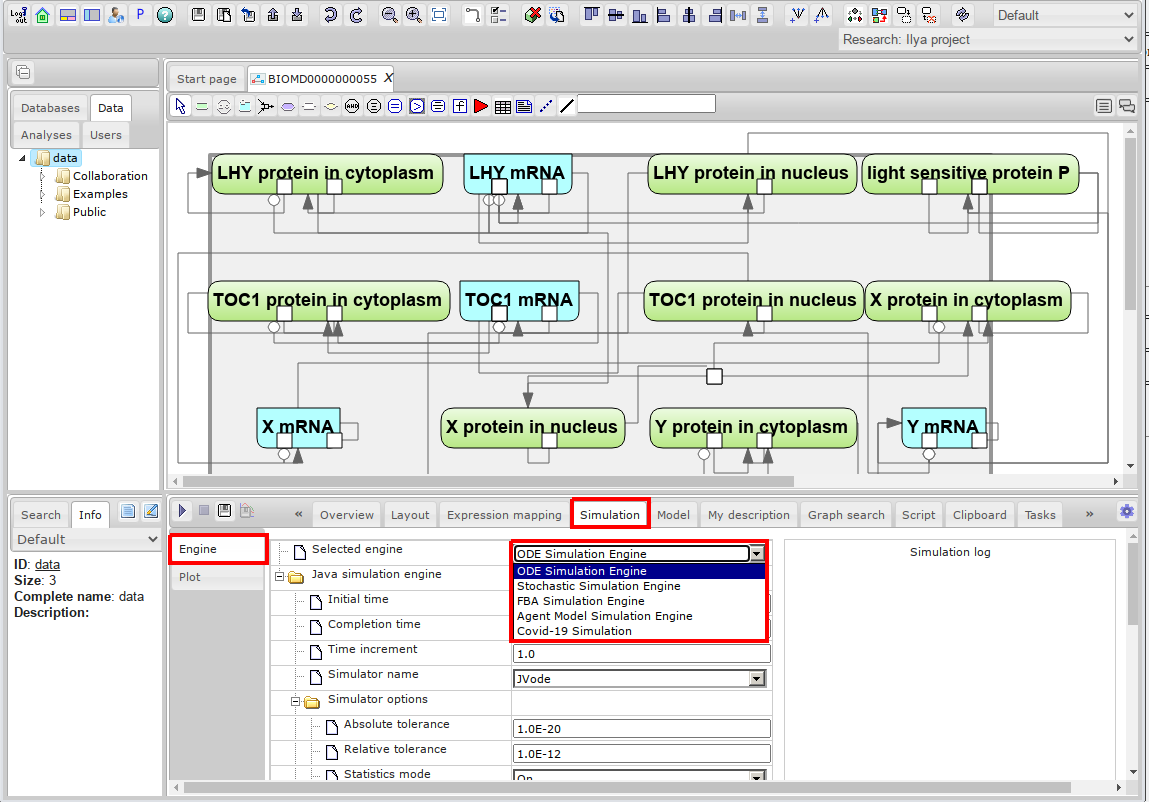
- Select one of the available simulators or use the default one:
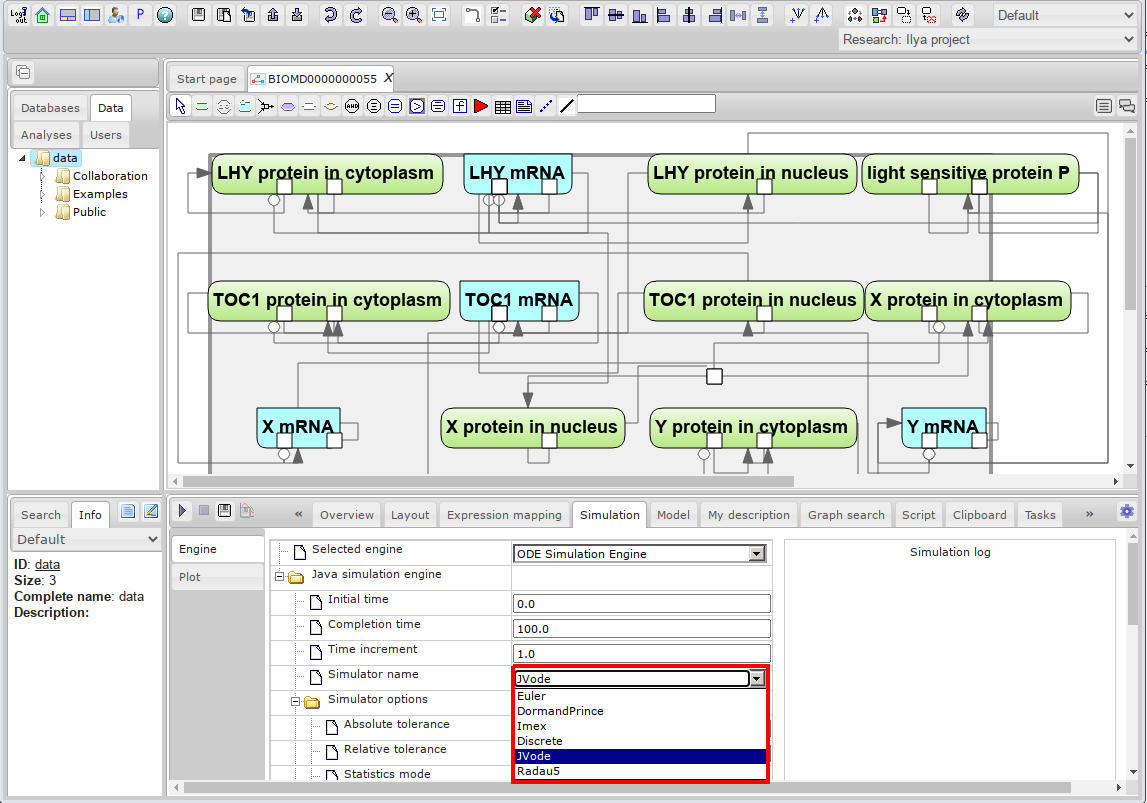
- Set simulation options:
- Initial time - initial value of the model time. It is strongly recommended to use only the value 0 here.
- Completion time - the point in model time when the simulation ends.
- Time increment - the gap between the time points at which the simulation results will be written.
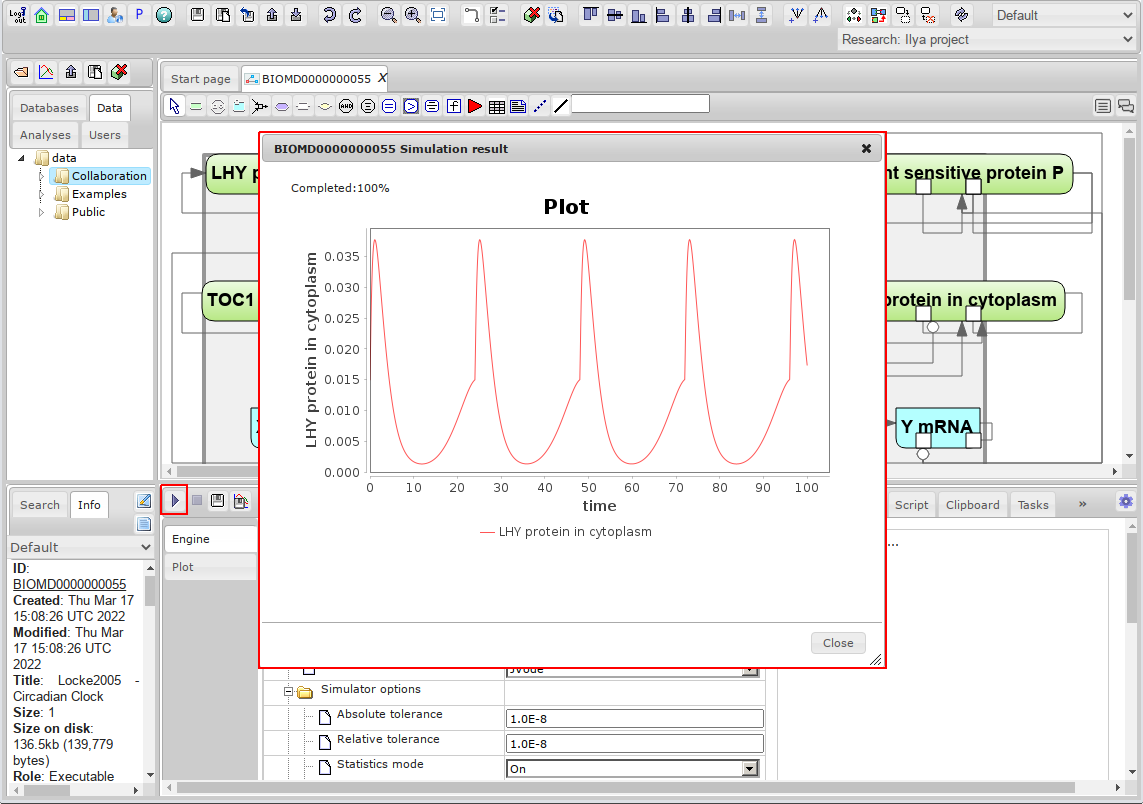
- Click on the Simulate button
 . The window that opens will show the results of simulation of the variables selected to plot.
. The window that opens will show the results of simulation of the variables selected to plot.