Difference between revisions of "Parameter identifiability example"
From BioUML platform
| Line 11: | Line 11: | ||
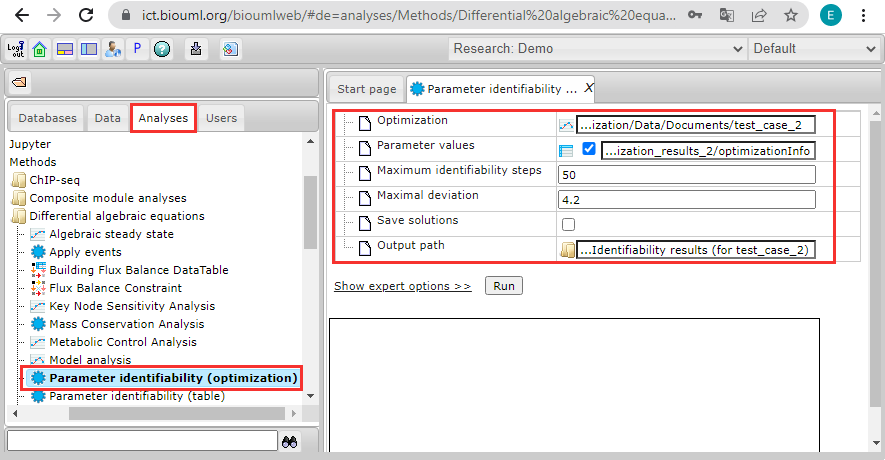
<h3>Parameter identifiability (optimization)</h3> | <h3>Parameter identifiability (optimization)</h3> | ||
| − | <br>[[File:parameter_identifiability_example_01.png]] | + | <br>[[File:parameter_identifiability_example_01.png]]<br> |
Ready analysis results can be found in the folder: | Ready analysis results can be found in the folder: | ||
Revision as of 15:36, 16 March 2022
Identifiability analysis infers how well the model parameters are approximated by the amount and quality of experimental data [1,2].
Contents[hide] |
Reproducing a test case in BioUML
To reproduce a test case below in the BioUML workbench, go to the Analyses tab in the navigation pane and follow to analyses > Methods > Differential algebraic equations.
Identifiability analysis can be run in two ways:
- to use a pre-created optimization document, double click on Parameter identifiability (optimization);
- for auto-generation of an optimization document using the given settings, double click on Parameter identifiability (table).
Parameter identifiability (optimization)
Ready analysis results can be found in the folder:
data/Examples/DAE models/Data/Parameter Identifiability
Parameter identifiability (table)
References
- Raue A, Kreutz C, Maiwald T, Bachmann J, Schilling M, Klingmüller U, Timmer J (2009) Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics, 25(15):1923–1929.
- Raue A, Becker V, Klingmüller U, Timmer J (2010) Identifiability and observability analysis for experimental design in nonlinear dynamical models. Chaos, 20(4):045105.