Difference between revisions of "Find common effectors in networks (GeneWays) (workflow)"
(GeneXplain -> geneXplain) |
(Adding provider-specific categories) |
||
| Line 27: | Line 27: | ||
[[Category:Workflows]] | [[Category:Workflows]] | ||
[[Category:Autogenerated pages]] | [[Category:Autogenerated pages]] | ||
| + | [[Category:geneXplain workflows]] | ||
Revision as of 11:09, 19 April 2013
- Workflow title
- Find common effectors in networks (GeneWays)
- Provider
- geneXplain GmbH
Workflow overview
Description
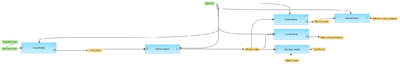
This workflow is designed to find effector molecules downstream of an input list of genes. Input file is any gene or protein table.
At the first step, the input table is converted into a table with Entrez Gene IDs.
At the next step the Entrez genes are subjected to Effector search in the GeneWays network. For each potential master regulator, FDR, Score, and Z-score are calculated.
The results are filtered by Z_Score>1 and Score>0.2 to select statistically significant Effector nodes.
Further, the filtered Effector molecules are converted to both Ensembl Gene IDs and a list of UniProt protein IDs.
The table with Ensembl Gene Ids is annotated with additional information, gene description and gene symbols.
Finally, the filtered Effector nodes table is sorted by Score and networks for the three top effectors are visualized as diagrams in the hierarchical layout.
Parameters
- Input gene set
- Species
- Results folder