Difference between revisions of "GTRD"
From BioUML platform
Ivan Yevshin (Talk | contribs) |
Ivan Yevshin (Talk | contribs) |
||
| Line 5: | Line 5: | ||
==Database statistics== | ==Database statistics== | ||
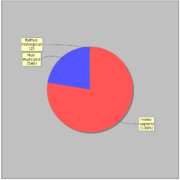
| − | GTRD uses 2417 ChIP-seq experiments for 470 distinct sequence specific transcription factors. | + | GTRD uses 2417 ChIP-seq experiments for 470 distinct sequence specific transcription factors. [[File:gtrd_exp_by_specie.png|thumb|ChIP-seq experiments by species]] |
| − | [[File:gtrd_exp_by_specie.png|ChIP-seq experiments by species]] | + | |
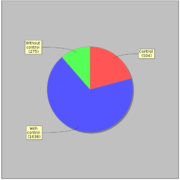
| − | Most of ChIP-seq experiments (1638) have corresponding control experiment. | + | Most of ChIP-seq experiments (1638) have corresponding control experiment. [[File:gtrd_exp_control.png|thumb|Control experiments]] |
| − | [[File:gtrd_exp_control.png|thumb|Control experiments]] | + | |
| + | In average each transcription factor is measured in 4.07 ChIP-seq experiments, but 284 (60%) transcription factors measured only in only one experiment. | ||
| + | The ten most studied transcription factors listed bellow: | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Transcription Factor !! Number of ChIP-seq experiments | ||
| + | |- | ||
| + | | CTCF || 195 | ||
| + | |- | ||
| + | | c-Myc || 45 | ||
| + | |- | ||
| + | | ERα || 44 | ||
| + | |- | ||
| + | | NRSF || 37 | ||
| + | |- | ||
| + | | C/EBPβ || 37 | ||
| + | |- | ||
| + | | GATA-1 || 33 | ||
| + | |- | ||
| + | | NF-κB p65 || 30 | ||
| + | |- | ||
| + | | Max || 30 | ||
| + | |- | ||
| + | | PU.1 || 29 | ||
| + | |- | ||
| + | | GR || 24 | ||
| + | |} | ||
| + | Transcription Factor ChIP-seq experiments count | ||
Revision as of 17:45, 1 July 2013
GTRD (Gene Transcription Regulation Database) is a database of transcription factor binding sites identified from ChIP-seq experiments. GTRD identifies binding sites from ChIP-seq experiments available from literature, GEO, SRA and ENCODE databases.
The web interface to GTRD is freely available at http://192.168.199.241/bioumlweb/#anonymous=true&perspective=GTRD.
Database statistics
GTRD uses 2417 ChIP-seq experiments for 470 distinct sequence specific transcription factors. Most of ChIP-seq experiments (1638) have corresponding control experiment.In average each transcription factor is measured in 4.07 ChIP-seq experiments, but 284 (60%) transcription factors measured only in only one experiment. The ten most studied transcription factors listed bellow:
| Transcription Factor | Number of ChIP-seq experiments |
|---|---|
| CTCF | 195 |
| c-Myc | 45 |
| ERα | 44 |
| NRSF | 37 |
| C/EBPβ | 37 |
| GATA-1 | 33 |
| NF-κB p65 | 30 |
| Max | 30 |
| PU.1 | 29 |
| GR | 24 |
Transcription Factor ChIP-seq experiments count