Difference between revisions of "GTRD"
Ivan Yevshin (Talk | contribs) |
Ivan Yevshin (Talk | contribs) |
||
| Line 147: | Line 147: | ||
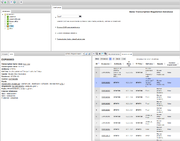
[[File:gtrd_search.png|thumb|Search GTRD]] | [[File:gtrd_search.png|thumb|Search GTRD]] | ||
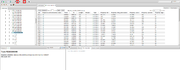
To search for stat transcription factors enter stat* in the search field and press enter, the list of matching ChIP-seq experiments will appear in the 'Search result' tab. Select one of them to view detailed ChIP-seq experiment information in the 'Info' tab. The 'Info' tab also provides links to the experiment data: reads, alignments and peaks. To view peaks or alignments click the link in the ChIP-seq experiment 'Info' tab, the track will be opened as table. This track can be exported by pressing 'Export' button on the toolbar, or opened in the 'Genome browser' by pressing 'Open as track' button on the toolbar. | To search for stat transcription factors enter stat* in the search field and press enter, the list of matching ChIP-seq experiments will appear in the 'Search result' tab. Select one of them to view detailed ChIP-seq experiment information in the 'Info' tab. The 'Info' tab also provides links to the experiment data: reads, alignments and peaks. To view peaks or alignments click the link in the ChIP-seq experiment 'Info' tab, the track will be opened as table. This track can be exported by pressing 'Export' button on the toolbar, or opened in the 'Genome browser' by pressing 'Open as track' button on the toolbar. | ||
| + | [[File:gtrd_open_peaks.png|thumb|Open ChIP-seq peaks]] | ||
For convenience some views are available as {{Type link|tree-table}}s. | For convenience some views are available as {{Type link|tree-table}}s. | ||
Revision as of 15:04, 2 July 2013
GTRD (Gene Transcription Regulation Database) is a database of transcription factor binding sites identified from ChIP-seq experiments. GTRD analyze freely avalable ChIP-seq experiments from literature, GEO, SRA and ENCODE databases.
The web interface to GTRD is available here.
Database statistics
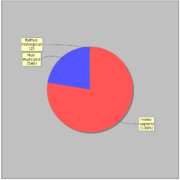
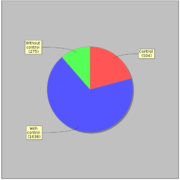
GTRD uses 2417 ChIP-seq experiments for 470 distinct sequence specific transcription factors. Most of ChIP-seq experiments (1638) have corresponding control experiment.General statistics:
| Object type | Total count | Per ChIP-seq experiment |
|---|---|---|
| ChIP-seq reads | 80.808E9 | 34.937E6 |
| Reads aligned | 58.848E9 | 25.675E6 |
| ChIP-seq peaks | 59.515E6 | 32899 |
In average each transcription factor is measured in 4.07 ChIP-seq experiments, but 284 (60%) transcription factors measured only in one experiment.
The ten most studied transcription factors listed bellow:
| Transcription Factor | Number of ChIP-seq experiments |
|---|---|
| CTCF | 195 |
| c-Myc | 45 |
| ERα | 44 |
| NRSF | 37 |
| C/EBPβ | 37 |
| GATA-1 | 33 |
| NF-κB p65 | 30 |
| Max | 30 |
| PU.1 | 29 |
| GR | 24 |
Database structure
The metadata concerning GTRD is stored in MySQL tables.
Each ChIP-seq experiment has a row in 'chip_experiments' table, which assigns id and stores basic information about experiment. 'chip_experiments' table has following structure:
| Column | Description | Example value |
|---|---|---|
| id | Unique experiment identifier | EXP000489 |
| antibody | Antibody used in chromatin immunoprecipitation | sc-345 |
| tfClassId | Id in TFClass[1] database of target transcription factor, NULL for control experiments | 6.2.1.0.1 |
| cell_line | Studied cell line | HeLa S3 |
| specie | Specie latin name | Homo sapiens |
| treatment | Cell treatment or conditions | IFN gamma |
| control_id | Id of control experiment, NULL for control experiments or experiments without control | EXP000490 |
The links to external databases stored in 'external_refs' table:
| Column | Description | Example values |
|---|---|---|
| id | Experiment identifier | EXP000489 |
| external_db | External database name | GEO or PUBMED or ENCODE or SRA |
| external_db_id | Identifier in external database | GSM320736 |
GTRD uses following object identifiers:
| Template | Object type | Example |
|---|---|---|
| EXPXXXXXX | ChIP-seq experiment | EXP000489 |
| READSXXXXXX | Collection of ChIP-seq reads | READS000770 |
| ALIGNSXXXXXX | Collection of read alignments | ALIGNS010001 |
| PEAKSXXXXXX | Collection of ChIP-seq peaks | PEAKS010000 |
The relationship between these objects is provided by 'hub' table:
| Column | Description | Example values |
|---|---|---|
| input | Input object identifier | READS000770 |
| input_type | Type of input object | ReadsGTRDType |
| output | Output object identifier | EXP000489 |
| output_type | Type of output object | ExperimentGTRDType |
ChIP-seq reads, alignments and peaks links to experiments with hub table in the following way:
| input | input_type | output | output_type |
|---|---|---|---|
| READS000770 | ReadsGTRDType | EXP000489 | ExperimentGTRDType |
| READS000771 | ReadsGTRDType | EXP000489 | ExperimentGTRDType |
| READS000772 | ReadsGTRDType | EXP000489 | ExperimentGTRDType |
| READS000773 | ReadsGTRDType | EXP000489 | ExperimentGTRDType |
| READS000774 | ReadsGTRDType | EXP000489 | ExperimentGTRDType |
| READS000775 | ReadsGTRDType | EXP000489 | ExperimentGTRDType |
| ALIGNS010001 | AlignmentsGTRDType | EXP000489 | ExperimentGTRDType |
| PEAKS010000 | PeaksGTRDType | EXP000489 | ExperimentGTRDType |
Web interface to database
| This page or section is a stub. Please add screenshots here! |
Web interface to GTRD is available at [2].
It provides capabilities for searching and browsing GTRD.
To search for stat transcription factors enter stat* in the search field and press enter, the list of matching ChIP-seq experiments will appear in the 'Search result' tab. Select one of them to view detailed ChIP-seq experiment information in the 'Info' tab. The 'Info' tab also provides links to the experiment data: reads, alignments and peaks. To view peaks or alignments click the link in the ChIP-seq experiment 'Info' tab, the track will be opened as table. This track can be exported by pressing 'Export' button on the toolbar, or opened in the 'Genome browser' by pressing 'Open as track' button on the toolbar.
For convenience some views are available as ![]() tree-tables.
tree-tables.