Difference between revisions of "Gene set enrichment analysis (Illumina probes) (workflow)"
(Automatic synchronization with BioUML) |
(Automatic synchronization with BioUML) |
||
| Line 6: | Line 6: | ||
[[File:Gene-set-enrichment-analysis-Illumina-probes-workflow-overview.png|400px]] | [[File:Gene-set-enrichment-analysis-Illumina-probes-workflow-overview.png|400px]] | ||
== Description == | == Description == | ||
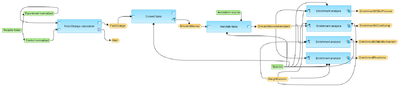
| − | This workflow is designed to perform Gene Set Enrichment Analysis, GSEA, as it is described at | + | This workflow is designed to perform Gene Set Enrichment Analysis, GSEA, as it is described at[http://www.broadinstitute.org/gsea/index.jsp http://www.broadinstitute.org/gsea/index.jsp]. As input, the normalized data with Illumina probeset IDs can be submitted. |
| − | Such normalized files are resulting from the “Normalize data” procedure under | + | Such normalized files are resulting from the “Normalize data” procedure under “analyses/Methods/Data normalization/Normalize Illumina experiment and control”. |
| − | + | First, the input files are subjected to fold-change calculation. The table with probeset Ids and calculated fold change values is converted into a table with Ensembl Gene Ids. | |
| − | + | At the next step, the Ensembl genes are annotated with additional information, gene description and gene symbols. | |
| + | |||
| + | Finally the annotated Ensembl genes are subjected to GSEA. Enrichment analysis is done in parallel by the following ontologies: GO biological processes, GO cellular components, GO molecular functions and by the Reactome pathways. | ||
| + | |||
| + | For each ontological item several parameters are calculated, including nominal p-value, ES, NES, as well as hit names, the link to the corresponding ontological term, and the link to open a visualization plot. | ||
| + | |||
| + | |||
== Parameters == | == Parameters == | ||
Revision as of 11:49, 30 July 2013
- Workflow title
- Gene set enrichment analysis (Illumina probes)
- Provider
- geneXplain GmbH
Workflow overview
Description
This workflow is designed to perform Gene Set Enrichment Analysis, GSEA, as it is described athttp://www.broadinstitute.org/gsea/index.jsp. As input, the normalized data with Illumina probeset IDs can be submitted.
Such normalized files are resulting from the “Normalize data” procedure under “analyses/Methods/Data normalization/Normalize Illumina experiment and control”.
First, the input files are subjected to fold-change calculation. The table with probeset Ids and calculated fold change values is converted into a table with Ensembl Gene Ids.
At the next step, the Ensembl genes are annotated with additional information, gene description and gene symbols.
Finally the annotated Ensembl genes are subjected to GSEA. Enrichment analysis is done in parallel by the following ontologies: GO biological processes, GO cellular components, GO molecular functions and by the Reactome pathways.
For each ontological item several parameters are calculated, including nominal p-value, ES, NES, as well as hit names, the link to the corresponding ontological term, and the link to open a visualization plot.
Parameters
- Experiment normalized
- Control normalized
- Species
- Results folder