Difference between revisions of "GTRD"
(→Repository structure) |
|||
| Line 6: | Line 6: | ||
* [http://genome.ucsc.edu/ENCODE/ ENCODE]. | * [http://genome.ucsc.edu/ENCODE/ ENCODE]. | ||
| − | Initial raw data were uniformly processed using specially developed workflow (pipeline) for BioUML platform: | + | Initial ChIP-seq and DNase-seq raw data were uniformly processed using specially developed workflow (pipeline) for BioUML platform. |
| + | ChIP-seq processing pipeline: | ||
* sequenced reads were aligned to reference genome using Bowtie2; | * sequenced reads were aligned to reference genome using Bowtie2; | ||
| − | * peaks were identified using MACS, SISSR, GEM and PICS peak callers | + | * peaks were identified using MACS, SISSR, GEM and PICS peak callers; |
| − | * peaks computed for the same TF and peak calling method, but different experiment conditions (e.g., cell line, treatment, etc.) were joined into clusters | + | * peaks computed for the same TF and peak calling method, but different experiment conditions (e.g., cell line, treatment, etc.) were joined into clusters; |
| − | * clusters for the same TF revealed by different peak calling methods were joined into metaclusters | + | * clusters for the same TF revealed by different peak calling methods were joined into metaclusters. |
| + | DNase-seq processing pipeline: | ||
| + | * sequenced reads were aligned to reference genome using Bowtie2; | ||
| + | * regions of open chromatin were identified using MACS2 and Hotspot2; | ||
| + | * to reveal de novo putative protein-DNA interactions digital genomic footprinting tool Wellington was used. | ||
[[GTRD_Workflow|Learn more about GTRD build process]] | [[GTRD_Workflow|Learn more about GTRD build process]] | ||
| Line 16: | Line 21: | ||
GTRD database is freely available for non-commercial organizations. | GTRD database is freely available for non-commercial organizations. | ||
| − | ==Database statistics== | + | ==Database statistics (18.06 version)== |
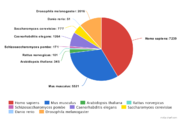
| − | GTRD uses | + | GTRD uses 17485 ChIP-seq experiments correspond to 2399 uniprot IDs. [[File:GTRD statistics-by-species3.png|thumb|ChIP-seq experiments by species]] |
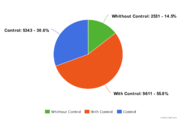
| − | Most of ChIP-seq experiments ( | + | Most of ChIP-seq experiments (79.2%) have corresponding control experiment. [[File:GTRD statistics-control3.png|thumb|Control experiments]] |
General statistics: | General statistics: | ||
| Line 26: | Line 31: | ||
! Object type !! Total count !! Per ChIP-seq experiment | ! Object type !! Total count !! Per ChIP-seq experiment | ||
|- | |- | ||
| − | | ChIP-seq reads || | + | | ChIP-seq reads || 719.3 × 10<sup>9</sup> || 41.1 × 10<sup>6</sup> |
|- | |- | ||
| − | | Reads aligned || | + | | Reads aligned || 539.8 × 10<sup>9</sup> || 30.9 × 10<sup>6</sup> |
|- | |- | ||
| − | | ChIP-seq peaks || > | + | | ChIP-seq peaks || >1.1 × 10<sup>9</sup> || depends on peak caller |
|} | |} | ||
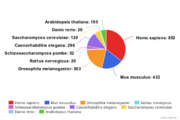
| − | In average each TF has been measured in | + | In average each TF has been measured in 7.28 ChIP-seq experiments, 52% of TFs have been measured in more than one experiment. [[File:GTRD tf-statistics-by-species3.png|thumb|Transription factors by species]] |
The ten most studied transcription factors are listed bellow: | The ten most studied transcription factors are listed bellow: | ||
| Line 39: | Line 44: | ||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| − | ! Transcription Factor !! Number of ChIP-seq experiments | + | !uniprot ID !! Transcription Factor !! Species !! Number of ChIP-seq experiments |
|- | |- | ||
| − | | CTCF || | + | | P49711 || CTCF || Homo sapiens || 432 |
|- | |- | ||
| − | | | + | | P03372 || SR1 || Homo sapiens || 280 |
|- | |- | ||
| − | | | + | | P10275 || ANDR || Homo sapiens || 259 |
|- | |- | ||
| − | | | + | | Q61164 || CTCF || Mus musculus || 255 |
|- | |- | ||
| − | | | + | | P17433 || SPI1 || Mus musculus || 156 |
|- | |- | ||
| − | | | + | | P55317 || FOXA1 || Homo sapiens || 125 |
|- | |- | ||
| − | | | + | | P01106 || MYC || Homo sapiens || 122 |
|- | |- | ||
| − | | | + | | P28033 || CEBPB || Mus musculus || 100 |
|- | |- | ||
| − | | | + | | Q04206 || TF65 || Homo sapiens || 93 |
|- | |- | ||
| − | | | + | | P04150 || GCR || Homo sapiens || 90 |
|} | |} | ||
| + | |||
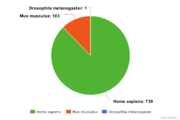
| + | GTRD contains 843 DNase-seq experiments. [[File:GTRD statistics-dnase-by-species3.png|thumb|DNase-seq experiments by species]] | ||
[[GTRD statistics|Detailed statistics]]. | [[GTRD statistics|Detailed statistics]]. | ||
| Line 67: | Line 74: | ||
The metadata concerning GTRD is stored in MySQL tables. | The metadata concerning GTRD is stored in MySQL tables. | ||
| − | Each ChIP-seq experiment has a row in 'chip_experiments' table, which assigns id and stores basic information about experiment. | + | Each ChIP-seq and DNase-seq experiment has a row in 'chip_experiments' or 'dnase-experiments' table, respectively, which assigns id and stores basic information about experiment. |
'chip_experiments' table has following structure: | 'chip_experiments' table has following structure: | ||
| Line 77: | Line 84: | ||
|- | |- | ||
| antibody || Antibody used in chromatin immunoprecipitation || sc-345 | | antibody || Antibody used in chromatin immunoprecipitation || sc-345 | ||
| − | |||
| − | |||
| − | |||
| − | |||
|- | |- | ||
| specie || Species latin name || Homo sapiens | | specie || Species latin name || Homo sapiens | ||
| Line 87: | Line 90: | ||
|- | |- | ||
| control_id || Id of control experiment, NULL for control experiments or experiments without control || EXP000490 | | control_id || Id of control experiment, NULL for control experiments or experiments without control || EXP000490 | ||
| + | |- | ||
| + | | cell_id || Studied cell line ID || 423 | ||
| + | |- | ||
| + | | experiment_type || || unspecified | ||
| + | |- | ||
| + | | tf_uniprot_id || Protein id in Uniprot database || P42224 | ||
| + | |} | ||
| + | |||
| + | 'dnase_experiments' table has following structure: | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Column !! Description || Example value | ||
| + | |- | ||
| + | | id || Unique experiment identifier || DEXP000044 | ||
| + | |- | ||
| + | | organism || Species latin name || Homo sapiens | ||
| + | |- | ||
| + | | treatment || Cell treatment or conditions || 100nM dexamethasone (CHEBI:41879) for 6 hour | ||
| + | |- | ||
| + | | cell_id || Studied cell line ID || 38 | ||
|} | |} | ||
| Line 111: | Line 135: | ||
| READSXXXXXX || Collection of ChIP-seq reads || READS000770 | | READSXXXXXX || Collection of ChIP-seq reads || READS000770 | ||
|- | |- | ||
| − | | ALIGNSXXXXXX || Collection of | + | | ALIGNSXXXXXX || Collection of ChIP-seq reads alignments || ALIGNS010001 |
|- | |- | ||
| − | | PEAKSXXXXXX || Collection of | + | | PEAKSXXXXXX || Collection of DNase-seq peaks || PEAKS010000 |
| + | |- | ||
| + | | DEXPXXXXXX || DNase-seq experiment || DEXP000044 | ||
| + | |- | ||
| + | | DREADSXXXXXX || Collection of DNase-seq reads || DREADS000232 | ||
| + | |- | ||
| + | | DALIGNSXXXXXX || Collection of DNase-seq reads alignments || DALIGNS000044 | ||
| + | |- | ||
| + | | DPEAKSXXXXXX || Collection of DNase-seq peaks || DPEAKS000044 | ||
|} | |} | ||
| Line 129: | Line 161: | ||
| output_type || Type of output object || ExperimentGTRDType | | output_type || Type of output object || ExperimentGTRDType | ||
|} | |} | ||
| − | ChIP-seq reads, alignments and peaks links to experiments with hub table in the following way: | + | ChIP-seq and DNase-seq reads, alignments and peaks links to experiments with hub table in the following way: |
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| − | ! input !! input_type !! output !! output_type | + | ! input !! input_type !! output !! output_type !! specie |
|- | |- | ||
| − | | READS000770 | + | | READS000770 || ReadsGTRDType || EXP000489 || ExperimentGTRDType || Homo sapiens |
|- | |- | ||
| − | | READS000771 | + | | READS000771 || ReadsGTRDType || EXP000489 || ExperimentGTRDType || Homo sapiens |
|- | |- | ||
| − | | READS000772 | + | | READS000772 || ReadsGTRDType || EXP000489 || ExperimentGTRDType || Homo sapiens |
|- | |- | ||
| − | | READS000773 | + | | READS000773 || ReadsGTRDType || EXP000489 || ExperimentGTRDType || Homo sapiens |
|- | |- | ||
| − | | READS000774 | + | | READS000774 || ReadsGTRDType || EXP000489 || ExperimentGTRDType || Homo sapiens |
|- | |- | ||
| − | | READS000775 | + | | READS000775 || ReadsGTRDType || EXP000489 || ExperimentGTRDType || Homo sapiens |
|- | |- | ||
| − | | | + | | ALIGNS033052 || AlignmentsGTRDType || EXP000489 || ExperimentGTRDType || Homo sapiens |
|- | |- | ||
| − | | | + | | PEAKS033050 || PeaksGTRDType || EXP000489 || ExperimentGTRDType || Homo sapiens |
| + | |- | ||
| + | | DALIGNS000044 || DNaseAlignments || DEXP000044 || DNaseExperiment || Homo sapiens | ||
| + | |- | ||
| + | | DPEAKS000044 || DNasePeaks || DEXP000044 || DNaseExperiment || Homo sapiens | ||
| + | |- | ||
| + | | DREADS000232 || DNaseReads || DEXP000044 || DNaseExperiment || Homo sapiens | ||
| + | |- | ||
| + | | DREADS000233 || DNaseReads || DEXP000044 || DNaseExperiment || Homo sapiens | ||
| + | |- | ||
| + | | DREADS000234 || DNaseReads || DEXP000044 || DNaseExperiment || Homo sapiens | ||
| + | |- | ||
| + | | DREADS000235 || DNaseReads || DEXP000044 || DNaseExperiment || Homo sapiens | ||
|} | |} | ||
| Line 158: | Line 202: | ||
===Start page=== | ===Start page=== | ||
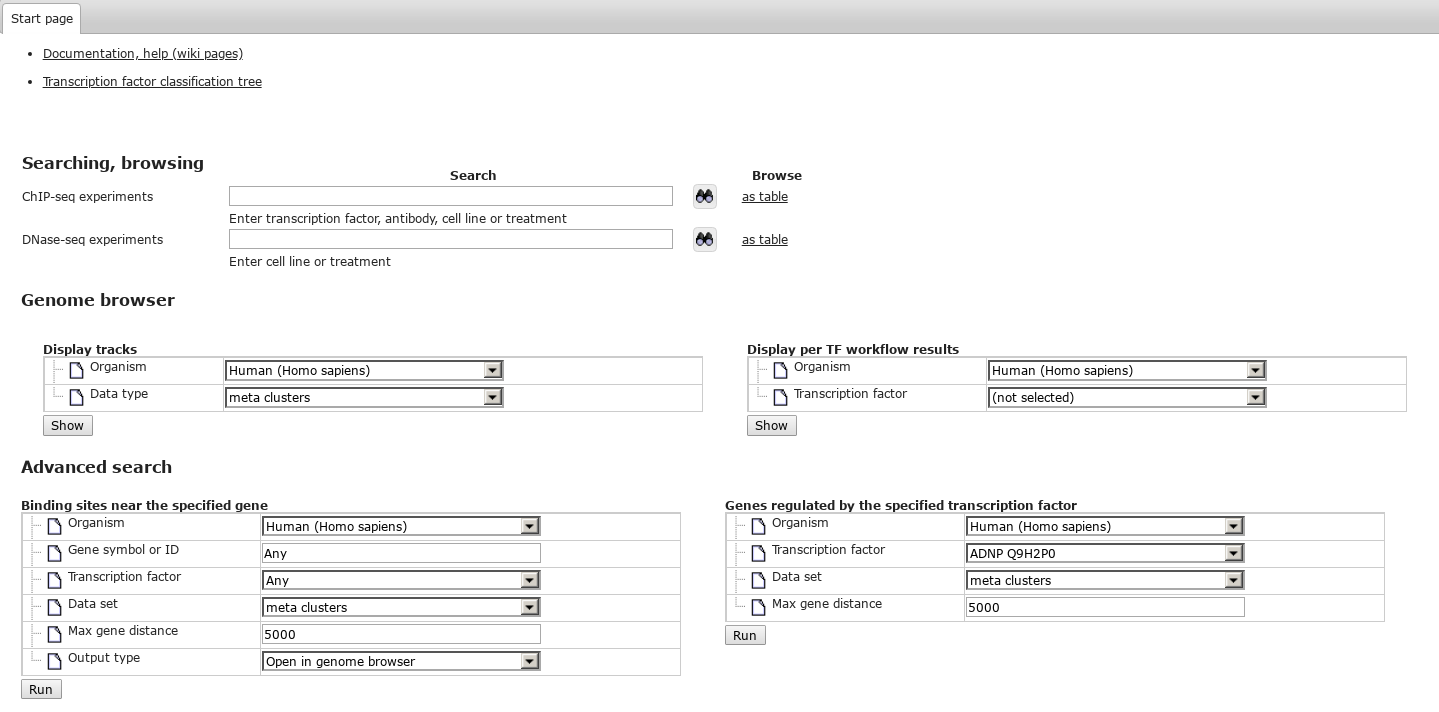
| − | Start page contains search box and the links to browse all experiments in the repository tree, | + | Start page contains search box and the links to browse all experiments in the repository tree, documentation, help (wiki pages) and transcription factor classification tree. |
| − | [[File: | + | [[File:gtrd_startpage3.png|GTRD start page]] |
===Search capabilities=== | ===Search capabilities=== | ||
| − | [[File: | + | [[File:gtrd_search3.png|thumb|Search GTRD]] |
[[File:gtrd_open_peaks.png|thumb|Open ChIP-seq peaks]] | [[File:gtrd_open_peaks.png|thumb|Open ChIP-seq peaks]] | ||
The ChIP-seq experiments contained in GTRD can be queried from the search box on the Start page. | The ChIP-seq experiments contained in GTRD can be queried from the search box on the Start page. | ||
GTRD uses [http://en.wikipedia.org/wiki/Lucene Lucene] engine for indexing and quering ChIP-seq experiments that provides rich [http://lucene.apache.org/core/2_9_4/queryparsersyntax.html syntax] for searching. | GTRD uses [http://en.wikipedia.org/wiki/Lucene Lucene] engine for indexing and quering ChIP-seq experiments that provides rich [http://lucene.apache.org/core/2_9_4/queryparsersyntax.html syntax] for searching. | ||
| − | The search can be performed by transcription factor(name or | + | The search can be performed by transcription factor(name or uniprot ID), cell line, antibody or treatment/conditions. |
For example, to search for STAT transcription factors enter stat* in the search field and press enter. | For example, to search for STAT transcription factors enter stat* in the search field and press enter. | ||
| Line 183: | Line 227: | ||
GTRD is organized in hierarchical [[Repository]]. | GTRD is organized in hierarchical [[Repository]]. | ||
| − | [[File: | + | [[File:gtrd_repository3.png|GTRD repository]] |
The GTRD/Data folder contains following items: | The GTRD/Data folder contains following items: | ||
| − | *experiments - | + | *DNase experiments - |
| − | + | ||
*alignments - ChIP-seq read alignments | *alignments - ChIP-seq read alignments | ||
| − | *peaks - ChIP-seq peaks identified by | + | *clusters - |
| + | *experiments - ChIP-seq experiments metainformation | ||
| + | *generic - | ||
| + | *peaks - ChIP-seq and DNase-seq peaks identified by peak callers | ||
| + | *views - TFClass centric view of GTRD data avalable as {{Type link|tree-table}}s | ||
GTRD/Dictionaries/classification is [http://www.edgar-wingender.de/huTF_classification.html TFClass] classification tree used by GTRD to reference transcription factors. | GTRD/Dictionaries/classification is [http://www.edgar-wingender.de/huTF_classification.html TFClass] classification tree used by GTRD to reference transcription factors. | ||
| + | GTRD/Dictionaries/cells is a collection of cells and tissues used by GTRD | ||
Revision as of 14:17, 22 October 2018
Gene Transcription Regulation Database (GTRD) is a database of transcription factor binding sites identified from ChIP-seq experiments that were systematically collected and uniformly processed using special workflow (pipeline) for BioUML platform. Raw ChIP-seq data and experiment information were collected from:
Initial ChIP-seq and DNase-seq raw data were uniformly processed using specially developed workflow (pipeline) for BioUML platform. ChIP-seq processing pipeline:
- sequenced reads were aligned to reference genome using Bowtie2;
- peaks were identified using MACS, SISSR, GEM and PICS peak callers;
- peaks computed for the same TF and peak calling method, but different experiment conditions (e.g., cell line, treatment, etc.) were joined into clusters;
- clusters for the same TF revealed by different peak calling methods were joined into metaclusters.
DNase-seq processing pipeline:
- sequenced reads were aligned to reference genome using Bowtie2;
- regions of open chromatin were identified using MACS2 and Hotspot2;
- to reveal de novo putative protein-DNA interactions digital genomic footprinting tool Wellington was used.
Learn more about GTRD build process
GTRD database is freely available for non-commercial organizations.
Contents |
Database statistics (18.06 version)
GTRD uses 17485 ChIP-seq experiments correspond to 2399 uniprot IDs. Most of ChIP-seq experiments (79.2%) have corresponding control experiment.General statistics:
| Object type | Total count | Per ChIP-seq experiment |
|---|---|---|
| ChIP-seq reads | 719.3 × 109 | 41.1 × 106 |
| Reads aligned | 539.8 × 109 | 30.9 × 106 |
| ChIP-seq peaks | >1.1 × 109 | depends on peak caller |
The ten most studied transcription factors are listed bellow:
| uniprot ID | Transcription Factor | Species | Number of ChIP-seq experiments |
|---|---|---|---|
| P49711 | CTCF | Homo sapiens | 432 |
| P03372 | SR1 | Homo sapiens | 280 |
| P10275 | ANDR | Homo sapiens | 259 |
| Q61164 | CTCF | Mus musculus | 255 |
| P17433 | SPI1 | Mus musculus | 156 |
| P55317 | FOXA1 | Homo sapiens | 125 |
| P01106 | MYC | Homo sapiens | 122 |
| P28033 | CEBPB | Mus musculus | 100 |
| Q04206 | TF65 | Homo sapiens | 93 |
| P04150 | GCR | Homo sapiens | 90 |
Database structure
The metadata concerning GTRD is stored in MySQL tables.
Each ChIP-seq and DNase-seq experiment has a row in 'chip_experiments' or 'dnase-experiments' table, respectively, which assigns id and stores basic information about experiment. 'chip_experiments' table has following structure:
| Column | Description | Example value |
|---|---|---|
| id | Unique experiment identifier | EXP000489 |
| antibody | Antibody used in chromatin immunoprecipitation | sc-345 |
| specie | Species latin name | Homo sapiens |
| treatment | Cell treatment or conditions | IFN gamma |
| control_id | Id of control experiment, NULL for control experiments or experiments without control | EXP000490 |
| cell_id | Studied cell line ID | 423 |
| experiment_type | unspecified | |
| tf_uniprot_id | Protein id in Uniprot database | P42224 |
'dnase_experiments' table has following structure:
| Column | Description | Example value |
|---|---|---|
| id | Unique experiment identifier | DEXP000044 |
| organism | Species latin name | Homo sapiens |
| treatment | Cell treatment or conditions | 100nM dexamethasone (CHEBI:41879) for 6 hour |
| cell_id | Studied cell line ID | 38 |
The links to external databases stored in 'external_refs' table:
| Column | Description | Example values |
|---|---|---|
| id | Experiment identifier | EXP000489 |
| external_db | External database name | GEO or PUBMED or ENCODE or SRA |
| external_db_id | Identifier in external database | GSM320736 |
GTRD uses following object identifiers:
| Template | Object type | Example |
|---|---|---|
| EXPXXXXXX | ChIP-seq experiment | EXP000489 |
| READSXXXXXX | Collection of ChIP-seq reads | READS000770 |
| ALIGNSXXXXXX | Collection of ChIP-seq reads alignments | ALIGNS010001 |
| PEAKSXXXXXX | Collection of DNase-seq peaks | PEAKS010000 |
| DEXPXXXXXX | DNase-seq experiment | DEXP000044 |
| DREADSXXXXXX | Collection of DNase-seq reads | DREADS000232 |
| DALIGNSXXXXXX | Collection of DNase-seq reads alignments | DALIGNS000044 |
| DPEAKSXXXXXX | Collection of DNase-seq peaks | DPEAKS000044 |
The relationship between these objects is provided by 'hub' table:
| Column | Description | Example values |
|---|---|---|
| input | Input object identifier | READS000770 |
| input_type | Type of input object | ReadsGTRDType |
| output | Output object identifier | EXP000489 |
| output_type | Type of output object | ExperimentGTRDType |
ChIP-seq and DNase-seq reads, alignments and peaks links to experiments with hub table in the following way:
| input | input_type | output | output_type | specie |
|---|---|---|---|---|
| READS000770 | ReadsGTRDType | EXP000489 | ExperimentGTRDType | Homo sapiens |
| READS000771 | ReadsGTRDType | EXP000489 | ExperimentGTRDType | Homo sapiens |
| READS000772 | ReadsGTRDType | EXP000489 | ExperimentGTRDType | Homo sapiens |
| READS000773 | ReadsGTRDType | EXP000489 | ExperimentGTRDType | Homo sapiens |
| READS000774 | ReadsGTRDType | EXP000489 | ExperimentGTRDType | Homo sapiens |
| READS000775 | ReadsGTRDType | EXP000489 | ExperimentGTRDType | Homo sapiens |
| ALIGNS033052 | AlignmentsGTRDType | EXP000489 | ExperimentGTRDType | Homo sapiens |
| PEAKS033050 | PeaksGTRDType | EXP000489 | ExperimentGTRDType | Homo sapiens |
| DALIGNS000044 | DNaseAlignments | DEXP000044 | DNaseExperiment | Homo sapiens |
| DPEAKS000044 | DNasePeaks | DEXP000044 | DNaseExperiment | Homo sapiens |
| DREADS000232 | DNaseReads | DEXP000044 | DNaseExperiment | Homo sapiens |
| DREADS000233 | DNaseReads | DEXP000044 | DNaseExperiment | Homo sapiens |
| DREADS000234 | DNaseReads | DEXP000044 | DNaseExperiment | Homo sapiens |
| DREADS000235 | DNaseReads | DEXP000044 | DNaseExperiment | Homo sapiens |
Web interface
| This page or section is a stub. Please add screenshots here! |
Web interface to GTRD is available here. It provides capabilities for searching and browsing GTRD.
Start page
Start page contains search box and the links to browse all experiments in the repository tree, documentation, help (wiki pages) and transcription factor classification tree.
Search capabilities
The ChIP-seq experiments contained in GTRD can be queried from the search box on the Start page. GTRD uses Lucene engine for indexing and quering ChIP-seq experiments that provides rich syntax for searching. The search can be performed by transcription factor(name or uniprot ID), cell line, antibody or treatment/conditions.
For example, to search for STAT transcription factors enter stat* in the search field and press enter. You can restrict query to HeLa cells treated with interferon with following query:
tfTitle:stat* AND cellLine:hela AND treatment:IFN
The list of matching ChIP-seq experiments will appear in the 'Search result' tab. Select one of them to view detailed ChIP-seq experiment information in the information box. The information box also provides links to the experiment data: reads, alignments and peaks.
To view peaks or alignments click the link in the ChIP-seq experiment information box, the ![]() track will be opened as table. The
track will be opened as table. The ![]() track can be exported by pressing 'Export' button or opened in the genome browser by pressing 'Open as track' button in the general control panel.
track can be exported by pressing 'Export' button or opened in the genome browser by pressing 'Open as track' button in the general control panel.
Repository structure
GTRD is organized in hierarchical Repository.
The GTRD/Data folder contains following items:
- DNase experiments -
- alignments - ChIP-seq read alignments
- clusters -
- experiments - ChIP-seq experiments metainformation
- generic -
- peaks - ChIP-seq and DNase-seq peaks identified by peak callers
- views - TFClass centric view of GTRD data avalable as
 tree-tables
tree-tables
GTRD/Dictionaries/classification is TFClass classification tree used by GTRD to reference transcription factors. GTRD/Dictionaries/cells is a collection of cells and tissues used by GTRD