Mapping to ontologies for multiple gene sets (TRANSPATH(R)) (workflow)
- Workflow title
- Mapping to ontologies for multiple gene sets (TRANSPATH(R))
- Provider
- geneXplain GmbH
Workflow overview
Description
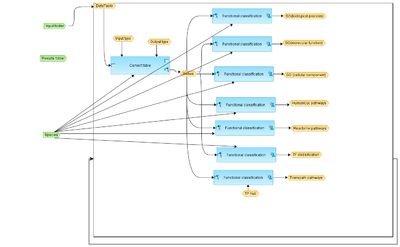
This workflow classifies multiple input gene sets using several ontologies and identifies categories that are over-represented in each of the input sets. The input is a folder containing several gene or protein tables.
In the first step, the first table from the input folder is converted into a gene table with Ensembl IDs.
In the second step, the table with Ensembl IDs is submitted to the Functional classification analysis, which is done in parallel using the following ontologies: GO biological processes, GO cellular components, GO molecular functions, Reactome pathways, HumanCyc pathways, TRANSPATH® pathways and TF classification.
The first and second steps are repeated for the second table from the input folder, and it is repeatedly performed for each table from the input folder.
As a result, a new folder is formed with several subfolders corresponding to each input table. Each subfolder contains the results of Functional classification. For each ontological category several parameters are calculated, including expected number of hits, actual number of hits, p-value as well as the names of genes falling into this category and the link to the corresponding ontological term.
This workflow is available together with a valid TRANSPATH® license.
Parameters
- Input folder
- Folder to get input tables from
- Species
- Results folder
- Folder to store results (will be created if not exists yet)