GTRD Workflow
ChIP-seq experiment information were collected in semi-automated way from literature, GEO and ENCODE.
Raw ChIP-seq data in the form of fastq and SRA files were fetched from ENCODE and SRA databases.
Sequenced reads were aligned using Bowtie2 aligner.
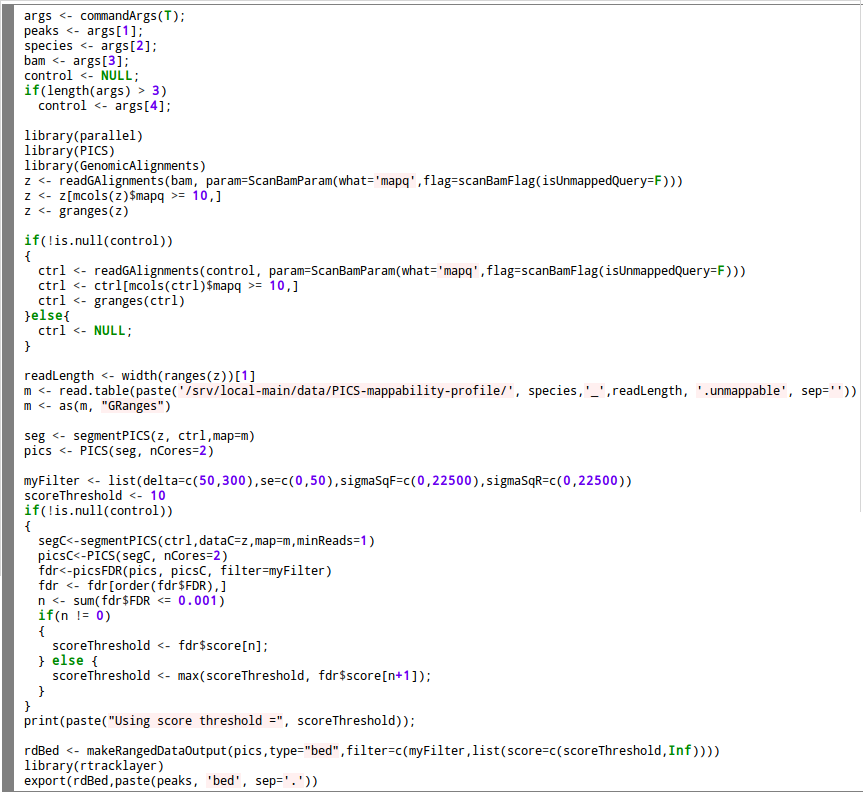
ChIP-seq peaks were called using 4 different methods: MACS SISSRS GEM and PICS.
Contents |
Bowtie2
We use bowtie2 version 2.2.3 for ChIP-seq read alignment to the reference genomes of human (GRCh38) and mouse (GRCm38).
Bowtie2 was run with following parameters:
bowtie2 -x $genome -U $fastq_files -p 8 --mm --seed 0
The resulting alignments were converted to bam files, then sorted and indexed using samtools version 1.0
MACS
MACS version 1.4.2 was used for peak calling with following parameters:
macs14 f BAM -g $species -n $peaks -t $alignment_bam
or if control experiment was available:
macs14 f BAM -g $species -n $peaks -t $alignment_bam -c $control_bam
SISSRS
SISSRS requires alignments in bed format, bam files were converted to bed files using bedtools version 2 by:
bamToBed -i $input_bam > $output_bed
Version 1.4 of SISSRS were used for peaks calling with following parameters:
sissrs.pl -i $alignment_bed -s 3000000000 -o $peaks.sissrs
or if control experiment was available:
sissrs.pl -i $alignment_bed -s 3000000000 -o $peaks.sissrs -b $control_bed
GEM
GEM version 2.5 was used with following parameters:
java -Xmx4G -XX:+UseSerialGC -jar /srv/local-main/tools/gem/gem.jar --d /srv/local-main/tools/gem/Read_Distribution_default.txt
--g /srv/local-main/tools/gem/$species.chrom.sizes --s 2000000000 --f SAM --t 1 --out $peaks --expt $bam
or if control experiment was available:
java -Xmx4G -XX:+UseSerialGC -jar /srv/local-main/tools/gem/gem.jar --d /srv/local-main/tools/gem/Read_Distribution_default.txt
--g /srv/local-main/tools/gem/$species.chrom.sizes --s 2000000000 --f SAM --t 1 --out $peaks --expt $bam --ctrl $control
For the large datasets -Xmx24G parameter was set.