Mapping to ontology - select a classification (2 Gene tables) (workflow)
- Workflow title
- Mapping to ontology - select a classification (2 Gene tables)
- Provider
- geneXplain GmbH
Workflow overview
Description
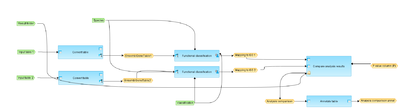
This workflow is designed to map each of the two input tables to one selected ontology classification, to identify term hits and to compare the results.
The input can be any gene or protein table. In the first step, the input tables are converted into two tables with Ensembl Gene IDs. These tables with Ensembl Gene IDs are subjected to a functional classification.
As result two mapped tables are stored and further compared via P-values. The comparison can help to reveal terms that show different enrichment across certain conditions. Output consists of a folder with sub-folder for each GO category.
The Analysis comparison annot table lists identifiers, annotation of identifiers, P-values for the first input set of genes and P-values for the second input set of genes (-log). The column Difference shows the absolute difference between two P-values. The columns Difference P-value and Difference FDR show the statistical significance of the absolute difference and upon sorting by one of these two columns on top you can see those ontology terms that are statistically most significantly different between two input gene sets. The comparison reveals GO terms that show different enrichment across the two input gene lists.
The Analysis comparison plot is a scatter plot of P-values on the log-scale together with the diagonal and the difference cutoffs at FDR < 0.05.
Parameters
- Input table 1
- Input table 2
- Species
- classification
- Result folder