Difference between revisions of "MGL"
(info transferred from the abstract on MGL) |
Tagir Valeev (Talk | contribs) m (links - →MGL structure) |
||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | '''MGL system (Molecular Genetics Library)''' is the object-oriented computer system for molecular genetic data management, analysis and visualization, which was designed in 1998 by a team of scientists at [http://www.bionet.nsc.ru/en/ Institute of Cytology and Genetics] (Siberian Branch of the [http://en.wikipedia.org/wiki/Russian_Academy_of_Sciences Russian Academy of Sciences]), Novosibirsk, Russia, for | |
* searching for and extracting information from molecular genetics databases; | * searching for and extracting information from molecular genetics databases; | ||
| − | * automated generation of nucleotide sequence samples of various gene components basing on semantic analysis of the EMBL FEATURE TABLE information and samples of promoters and transcription factor binding sites basing on the information from the EPD and TRRD databases, respectively; | + | * automated generation of nucleotide sequence samples of various gene components basing on semantic analysis of the EMBL FEATURE TABLE information and samples of promoters and transcription factor binding sites basing on the information from the {{EPD}} and [[TRRD]] databases, respectively; |
* nucleotide sequences analysis; and | * nucleotide sequences analysis; and | ||
* visualization of the database-contained information and the results of analyses performed. | * visualization of the database-contained information and the results of analyses performed. | ||
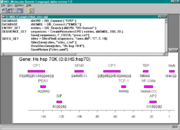
| − | [[File:MGLsystem. | + | [[File:MGLsystem.png|thumb|Operation of the system MGL. The upper window contains the text of the program; the lower, the map of the transcription factor binding sites found for a specified gene.]] |
The core of the MGL was the class library based on the idea that the set of classes corresponds directly to the main concepts of molecular genetic data. A specialized high-level object-oriented language allowed a user to generate samples and analyze nucleotide sequences in the automatic mode. The MGL system's user interface was designed for the MS Windows OS. The system had a demo version available online. | The core of the MGL was the class library based on the idea that the set of classes corresponds directly to the main concepts of molecular genetic data. A specialized high-level object-oriented language allowed a user to generate samples and analyze nucleotide sequences in the automatic mode. The MGL system's user interface was designed for the MS Windows OS. The system had a demo version available online. | ||
| Line 25: | Line 25: | ||
(5) for graphical representation of the data from databases and the results obtained. | (5) for graphical representation of the data from databases and the results obtained. | ||
| − | The library was realized in C++; in addition, sections 1, 2, and 5 were also realized in Java and used to develop the applications for Internet-based visualization of gene networks within the GeneNet database and transcription regulatory regions within the TRRD database. | + | The library was realized in C++; in addition, sections 1, 2, and 5 were also realized in Java and used to develop the applications for Internet-based visualization of gene networks within the [[GeneNet]] database and transcription regulatory regions within the [[TRRD]] database. |
'''Molecular Genetics Language (MGL)''' as a specialized high-level object-oriented language had special types corresponding to the basic molecular genetic notions (for example, SITE, SEQUENCE, SITE_SET, SEQUENCE_SET, etc.), the notions connected with database operation (DATABASE, ENTRY, ENTRY_SET, etc.), and corresponding to certain types of analysis (ALIGNMENT, PROFILE). | '''Molecular Genetics Language (MGL)''' as a specialized high-level object-oriented language had special types corresponding to the basic molecular genetic notions (for example, SITE, SEQUENCE, SITE_SET, SEQUENCE_SET, etc.), the notions connected with database operation (DATABASE, ENTRY, ENTRY_SET, etc.), and corresponding to certain types of analysis (ALIGNMENT, PROFILE). | ||
'''Graphical user interface''' was realized for the MS Windows OS and incorporated an editor for MGL programs, windows for viewing results in textual and graphical forms, and a set of dialogues for generating samples of nucleotide sequences and their analysis. | '''Graphical user interface''' was realized for the MS Windows OS and incorporated an editor for MGL programs, windows for viewing results in textual and graphical forms, and a set of dialogues for generating samples of nucleotide sequences and their analysis. | ||
| − | |||
==MGL possibilities== | ==MGL possibilities== | ||
| Line 47: | Line 46: | ||
MGL provided for automated generation of the nucleotide sequence samples. The samples of various genes components (promoters, introns, exons, splicing sites, polyadenylation sites, etc.) are generated based on semantic analysis of the FEATURE TABLE of the EMBL database. The semantic analysis includes checking the information for compliance with the basic principles of the eukaryotic gene organization. The system MGL provided the user with two modes of sample generation: (1) completely automatic and (2) allowing errors to be manually corrected. In the first mode, the system would try to correct the error in the gene structure description by itself based on the available knowledge; in the second case, the user would have to correct the gene structure description, and then the system would analyze it again. | MGL provided for automated generation of the nucleotide sequence samples. The samples of various genes components (promoters, introns, exons, splicing sites, polyadenylation sites, etc.) are generated based on semantic analysis of the FEATURE TABLE of the EMBL database. The semantic analysis includes checking the information for compliance with the basic principles of the eukaryotic gene organization. The system MGL provided the user with two modes of sample generation: (1) completely automatic and (2) allowing errors to be manually corrected. In the first mode, the system would try to correct the error in the gene structure description by itself based on the available knowledge; in the second case, the user would have to correct the gene structure description, and then the system would analyze it again. | ||
| − | While generating the samples of promoter regions and transcription factor binding sites, the system MGL used the EMBL database as a source of nucleotide sequences and the EPD and TRRD databases as a source of the data on location of the corresponding functional sites in these sequences. | + | While generating the samples of promoter regions and transcription factor binding sites, the system MGL used the EMBL database as a source of nucleotide sequences and the {{EPD}} and [[TRRD]] databases as a source of the data on location of the corresponding functional sites in these sequences. |
| − | The nucleotide sequence samples generated could be stored in different formats (EMBL-like, FASTA, PIR, etc.). In addition, the system MGL provided an easy conversion from one format to another. | + | The nucleotide sequence samples generated could be stored in different formats ([[EMBL format|EMBL-like]], [[Fasta format|FASTA]], PIR, etc.). In addition, the system MGL provided an easy conversion from one format to another. |
| Line 59: | Line 58: | ||
'''Visualization of the information from databases and the results obtained''' | '''Visualization of the information from databases and the results obtained''' | ||
| − | The system MGL also allowed the information from databases and the results obtained to be represented graphically. For example, the data from EMBL would be represented as a scheme of the structure of the gene and its functional sites; the data from TRANSFAC and TRRD, as graphical maps of gene regulatory regions. Various functional sites found as a result of the analysis performed could be also represented as a graphical map. | + | The system MGL also allowed the information from databases and the results obtained to be represented graphically. For example, the data from EMBL would be represented as a scheme of the structure of the gene and its functional sites; the data from {{TRANSFAC}} and [[TRRD]], as graphical maps of gene regulatory regions. Various functional sites found as a result of the analysis performed could be also represented as a graphical map. |
| Line 65: | Line 64: | ||
Automated recording of all the steps of operation was an important characteristic of the MGL system. For this purpose, the system would create a special file, serving as a working logbook. This file contained the dates of generation of the samples and performance of the analyses of nucleotide sequences, names of the functions and their options used, names of the files containing the samples generated and the results obtained, messages of the system in the course of operation, etc. This gave the user complete information on the process of automated sample generation and sequence analysis. | Automated recording of all the steps of operation was an important characteristic of the MGL system. For this purpose, the system would create a special file, serving as a working logbook. This file contained the dates of generation of the samples and performance of the analyses of nucleotide sequences, names of the functions and their options used, names of the files containing the samples generated and the results obtained, messages of the system in the course of operation, etc. This gave the user complete information on the process of automated sample generation and sequence analysis. | ||
| − | |||
==External links== | ==External links== | ||
*[http://www.bionet.nsc.ru/meeting/bgrs/thesis/125/ The abstract on MGL published for the International Conference on Bioinformatics of Genome Regulation and Structure 1998] | *[http://www.bionet.nsc.ru/meeting/bgrs/thesis/125/ The abstract on MGL published for the International Conference on Bioinformatics of Genome Regulation and Structure 1998] | ||
*[http://www.bionet.nsc.ru/en/ Website of Institute of Cytology and Genetics] | *[http://www.bionet.nsc.ru/en/ Website of Institute of Cytology and Genetics] | ||
| + | *[http://www.nsc.ru/en/ Website of the Siberian Branch of the Russian Academy of Sciences] | ||
| + | |||
| + | [[Category:BioUML history]] | ||
| + | [[Category:Software]] | ||
Latest revision as of 13:42, 2 July 2013
MGL system (Molecular Genetics Library) is the object-oriented computer system for molecular genetic data management, analysis and visualization, which was designed in 1998 by a team of scientists at Institute of Cytology and Genetics (Siberian Branch of the Russian Academy of Sciences), Novosibirsk, Russia, for
- searching for and extracting information from molecular genetics databases;
- automated generation of nucleotide sequence samples of various gene components basing on semantic analysis of the EMBL FEATURE TABLE information and samples of promoters and transcription factor binding sites basing on the information from the EPD and TRRD databases, respectively;
- nucleotide sequences analysis; and
- visualization of the database-contained information and the results of analyses performed.
The core of the MGL was the class library based on the idea that the set of classes corresponds directly to the main concepts of molecular genetic data. A specialized high-level object-oriented language allowed a user to generate samples and analyze nucleotide sequences in the automatic mode. The MGL system's user interface was designed for the MS Windows OS. The system had a demo version available online.
The work was supported by grants from the Russian Foundation for Basic Research (No.97-04-49740, 97-07-90309, 96-04-50006, 98-04-49479, 98-07-90126); Russian Ministry of Science and Technologies; Russian Human Genome Project; Russian Ministry of High Education.
[edit] MGL structure
The computer system MGL consisted of three sections: (1) an object-oriented class library; (2) a specialized high-level object-oriented language; and (3) a user interface.
The object-oriented class library included classes that could be divided into five major groups:
(1) the classes corresponding to the basic molecular genetic notions (Sequence, Site, GeneStructure, etc.);
(2) the classes for operation with databases;
(3) for generating samples of genomic sequences;
(4) for analyzing genomic sequences;
(5) for graphical representation of the data from databases and the results obtained.
The library was realized in C++; in addition, sections 1, 2, and 5 were also realized in Java and used to develop the applications for Internet-based visualization of gene networks within the GeneNet database and transcription regulatory regions within the TRRD database.
Molecular Genetics Language (MGL) as a specialized high-level object-oriented language had special types corresponding to the basic molecular genetic notions (for example, SITE, SEQUENCE, SITE_SET, SEQUENCE_SET, etc.), the notions connected with database operation (DATABASE, ENTRY, ENTRY_SET, etc.), and corresponding to certain types of analysis (ALIGNMENT, PROFILE).
Graphical user interface was realized for the MS Windows OS and incorporated an editor for MGL programs, windows for viewing results in textual and graphical forms, and a set of dialogues for generating samples of nucleotide sequences and their analysis.
[edit] MGL possibilities
Database access
Computer system MGL provided access (search for and extraction of information) to the databases
- accessible via the SRS (Sequence Retrieval System; Etzold, Argos, 1993) and
- installed on the user's computer (EMBL, ENZYME, EPD, PROSITE, SWISS-PROT, etc.).
In addition, the MGL systems supported operations with various samples generated from databases.
Automated generation of nucleotide sequence samples
MGL provided for automated generation of the nucleotide sequence samples. The samples of various genes components (promoters, introns, exons, splicing sites, polyadenylation sites, etc.) are generated based on semantic analysis of the FEATURE TABLE of the EMBL database. The semantic analysis includes checking the information for compliance with the basic principles of the eukaryotic gene organization. The system MGL provided the user with two modes of sample generation: (1) completely automatic and (2) allowing errors to be manually corrected. In the first mode, the system would try to correct the error in the gene structure description by itself based on the available knowledge; in the second case, the user would have to correct the gene structure description, and then the system would analyze it again.
While generating the samples of promoter regions and transcription factor binding sites, the system MGL used the EMBL database as a source of nucleotide sequences and the EPD and TRRD databases as a source of the data on location of the corresponding functional sites in these sequences.
The nucleotide sequence samples generated could be stored in different formats (EMBL-like, FASTA, PIR, etc.). In addition, the system MGL provided an easy conversion from one format to another.
Methods for nucleotide sequences analysis
The computer system MGL contained a wide range of tools for analysis of nucleotide sequences including calculation of nucleotide and oligonucleotide compositions; pairwise general and local alignments; rapid estimation of pairwise general alignment significance; calculation of the number of synonymous and nonsynonymous substitutions; analysis of leader sequences; calculation of similarity profiles for groups of functionally related sequences basing on their pairwise local alignments; search for transcription factor binding sites, etc.
Visualization of the information from databases and the results obtained
The system MGL also allowed the information from databases and the results obtained to be represented graphically. For example, the data from EMBL would be represented as a scheme of the structure of the gene and its functional sites; the data from TRANSFAC® Professional database and TRRD, as graphical maps of gene regulatory regions. Various functional sites found as a result of the analysis performed could be also represented as a graphical map.
Automated recording of operation
Automated recording of all the steps of operation was an important characteristic of the MGL system. For this purpose, the system would create a special file, serving as a working logbook. This file contained the dates of generation of the samples and performance of the analyses of nucleotide sequences, names of the functions and their options used, names of the files containing the samples generated and the results obtained, messages of the system in the course of operation, etc. This gave the user complete information on the process of automated sample generation and sequence analysis.